Pim-1 is an oncogene-encoded serine/threonine kinase. Originally identified in Maloney murine leukaemia, it is involved in several cellular functions associated with survival an proliferation which confers a selective advantage during tumorigenesis [1,2]. Given this implication, it has been used as a cancer drug target [3].
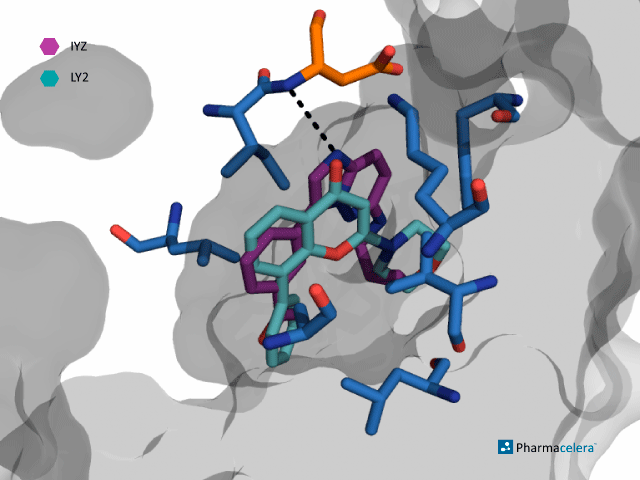
IYZ and LY2 are two bioactive inhibitors of Pim-1. The main described interactions between the protein and these molecules are hydrophobic. This can be appreciated in the reference molecule in the picture below (blue residues: Lys67, Asp186, Val52, Ile185, Leu44, Phe49, Leu174, Ala65), which also shows one hydrogen bond interaction (orange residue: Lys67).
Tools using hydrophobic parameters derived from solvation models, such as a quantum mechanical (QM) version of the MST continuum method used in PharmScreen, favours this type of ligand-target interactions.
The similarity-property principle suggests that analogous compounds will likely share similar biological properties. Indeed, defining the adequate properties that define the biological interactions are fundamental to explore similarity studies. In this case, hydrophobicity is an essential interaction to be considered when a ligand-based drug design process is performed.
Alignment
In order to verify it, we have aligned IYZ against LY2 using both traditional interaction fields and PharmScreen´s hydrophobic interaction fields and the results have been compared with the crystal structure.
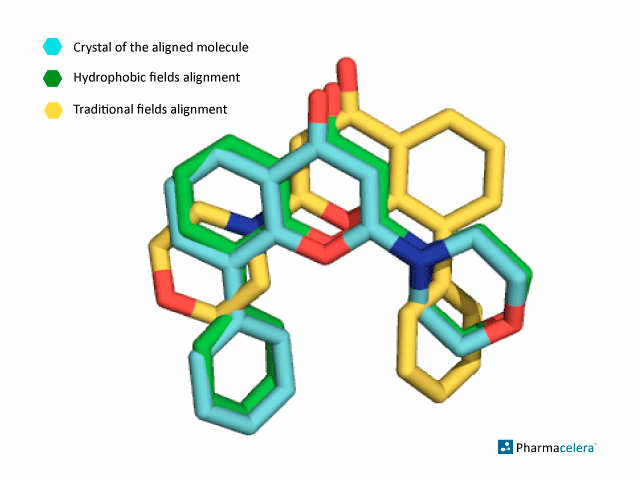
The picture below shows the alignment of both approaches with respect to the reference molecule in purple.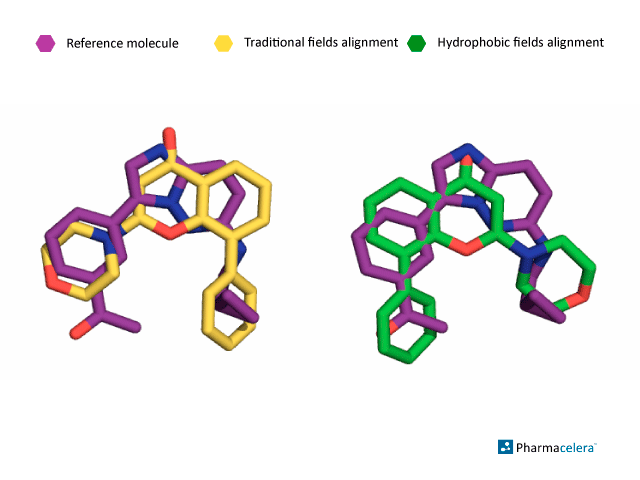 When comparing this with the crystallized molecule, the alignment performed considering traditional interaction fields misses the correct pose of the molecule, while PharmScreen is able to find the bioactive overlay using hydrophobic interaction fields, as shown in the picture below.
When comparing this with the crystallized molecule, the alignment performed considering traditional interaction fields misses the correct pose of the molecule, while PharmScreen is able to find the bioactive overlay using hydrophobic interaction fields, as shown in the picture below.
Hence, when searching for new potential hits in ligand-based in-silico approaches, it is crucial to use models for molecular alignment and similarity that use hydrophobic properties in situations in which hydrophobicity dominates the interaction between a ligand and a protein, as the one shown in this example.
[1] C. J. Saris, J. Domen, and A. Berns, “The pim-1 oncogene encodes two related protein-serine/threonine kinases by alternative initiation at AUG and CUG.,” EMBO J., vol. 10, no. 3, pp. 655–64, Mar. 1991.
[2] J. J. Gu, Z. Wang, R. Reeves, and N. S. Magnuson, “PIM1 phosphorylates and negatively regulates ASK1-mediated apoptosis.,” Oncogene, vol. 28, no. 48, pp. 4261–71, Dec. 2009.
[3] Y. Tursynbay, J. Zhang, Z. Li, T. Tokay, Z. Zhumadilov, D. Wu, and Y. Xie, “Pim-1 kinase as cancer drug target: An update.,” Biomed. reports, vol. 4, no. 2, pp. 140–146, Feb. 2016.