Fernando Martin
May 4th, 2023
An exponential growth of the accessible chemical space
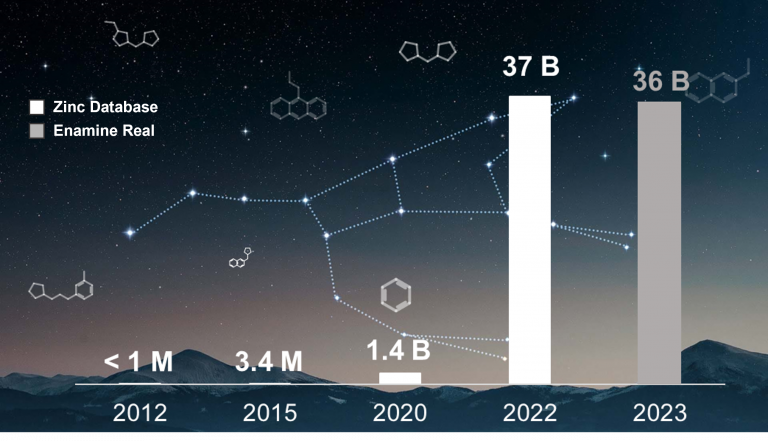
In the last years, there has been an exponential growth in commercial chemical libraries from millions to billions. To put some numbers: ZINC database has increased its size 37.000-fold since its 2012 version and Enamine REAL size is now over the 36 billion of compounds. Traditional computational chemistry approaches might not be usable anymore with these libraries due to computational and timing costs.
Enric Herrero, CTO at Pharmacelera, had the chance to talk about this topic at the Boston Area Group for Informatics and Modeling (BAGIM) last march. The presentation pointed out the need for novel tools that help exploring this novel ultra-large available chemical space and why current methods struggle to deal with it. During the presentation, different methods were listed, focusing in alternatives for ligand-based methods: brute-force (full enumeration), sampling methods and combinatorial search. Special emphasis was applied to the two latest, since they offer an alternative to screen larger chemical spaces than brute force using fewer computing resources. In this post we will quickly introduce approaches: MolPAL, based on sampling methods; and combinatorial search using 3D hydrophobic descriptors.
Sampling virtual screening powered by AI: MolPAL
This sampling method, based on David E. Graff paper, establishes that only a small subset (around 2.4%) of a full library must be evaluated with a slow screening method (i.e. docking or 3D ligand-based similarity) to obtain the same results than a brute-force method.
This is done in an iterative way, sampling first a random subset (~0.4%), evaluating the score with the slow method and then using the results to train a machine learning (ML) model. This model would be later applied to predict the score of all the ligands in the full library and select a new subset to be evaluated with the slow method.
To evaluate the capabilities of the method, our team has applied PharmScreen as scoring method to train a machine learning model. One of the main advantages observed when comparing MolPAL with brute-force screening (here, run PharmScreen for a full library), is the reduction in terms library storage (1B of compounds will suppose 44TB with brute force while MolPAL method will require only 96MB) as well as the computing speed.
An important aspect of sampling methods is that their performance will be linked to the speed ratio of the method used to mine the library vs the ML model training and prediction: the slowest or more computationally intensive the method is with respect to the ML training and prediction, the better in terms of speed gain.
Combinatorial search
Combinatorial search methods take advantage of the combinatorial chemistry concept, where the chemical space is explored using building block libraries and reaction information. By partitioning a reference molecule in fragments, screening software tools based on this method can explore libraries to find similar build blocks and enumerate only those compounds that are more similar. Since the reactivity of these building blocks is considered, one can easily reconstruct novel and synthesizable compounds that can be easily tested in the laboratory.
These methods can also take advantage of 3D information and the derived physicochemical properties when assessing the building block similarity. We have observed how the application of 3D hydrophobic molecular descriptors can help finding more diverse compounds with similar physicochemical properties than 2D methods.
Combinatorial search methods provide the best scalability among all the evaluated methods and, therefore, are a good alternative for the screening of multi-billion sized libraries. To put an example, using 3D methods as mentioned above and a library of 137K building blocks, we can explore a potential space of 31B of synthesizable molecules.
Conclusions
The exponential growth of the accessible chemical space is driving the generation of new methods that will help screening it. Contrary to full enumeration methods, that explore large libraries in exchange of using more computational resources, these new approaches can explore huger chemical libraries with more feasible hardware configurations.
Sampling methods, such as MolPAL, represent a good choice when screening large libraries using computing-demanding methods, such as docking or 3D ligand-based similarity. These methods are also interesting when storage capabilities suppose a problem.
Similarly, combinatorial search methods are a smart solution when screening ultra-large libraries, such as Enamine REAL. The use of building block libraries while considering their reactivity maximizes the synthesizability of novel compounds.
Pharmacelera is focused on offering novel solutions to explore this ultra-large chemical space. If you want to learn more about it, contact our team. They will inform you about our new services in this field and new technologies to come.