2019 was an exciting year for Pharmacelera and we look forward to 2020 with energy and commitment. Stay tuned for updates in forthcoming months! Let’s have a look to the 2019 summary!
Collaborations and Partnerships

This year has been very productive for us. For example, we have started a new collaboration with a top 10 pharmaceutical company. And we now have users and customers in Spain, France, Italy, Sweden, Germany, the United Kingdom and the United States.
In June, we also signed a partnership agreement with the Cambridge Crystallographic Data Center to explore the complementarity of our CADD tools and to apply machine learning for rational drug design.
In December, we became technology partners of KNIME. Now you will be able to access PharmScreen® through KNIME in early 2020. If you are using other Scientific Workflow Systems (SWS) rather than KNIME and you want to test PharmScreen®, do not hesitate to let us know and we will help you.
Finally, during this year, we have engaged with several public research institutions that apply PharmScreen® to identify novel hits targeting Parkinson, malaria, GPCRs and cancer through our academic program. Contact us if you are interested in applying our technology at your public research organization.
PharmScreen and software
We added new options to access our technology and expertise. You can now explore a richer chemical space using PharmScreen® through Software-as-a-Service (SaaS), on-premise yearly license, KNIME software modules or hiring our computational chemistry services. We will identify the option that fits you best to find hits with larger chemical diversity.
In terms of technology updates, we are happy to announce that we finalized a study about PharmScreen® complementing molecular docking tools such as Glide, Gold and rDock. In this study, we have observed that PharmScreen®’s hydrophobic similarity function significantly improves the results of structure-based drug design virtual screening in more than 40 diverse datasets. This feature is available as a software module. Let us know if you would like to become a beta tester for this module applied to other molecular docking packages.
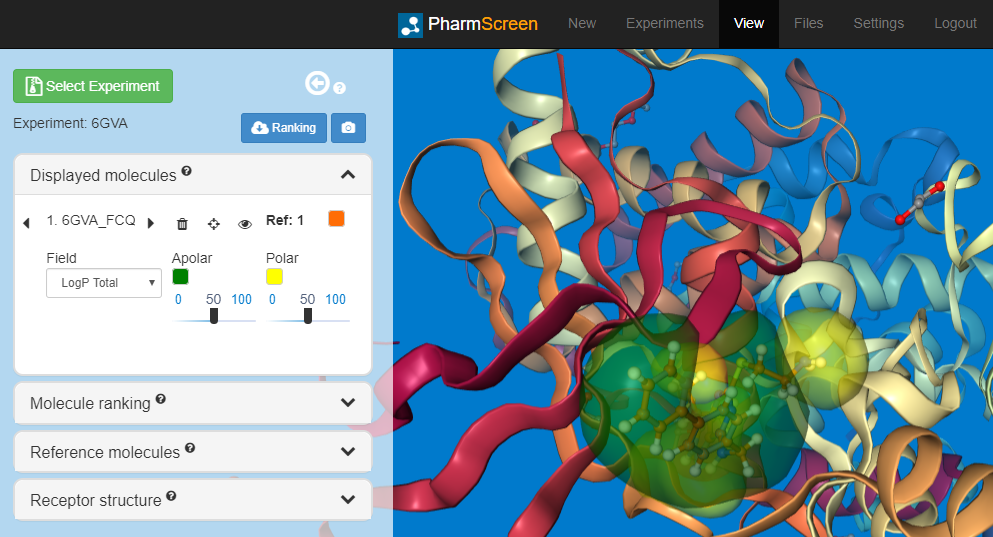
We have also deployed a new version of our Software-as-a-Service (SaaS) Graphical User Interface (GUI). Among other enhancements, you can now visualize the 3D hydrophobic interaction fields of ligands and its overlay within the target protein, as well as a description of the hydrogen bonds. We have also added to additional force fields, UFF and MMFF94s, to the existing ones, AM1 and RM1, to minimize the compounds of your selected library and reference molecule.
Together with your preferred library (your internal libraries, public libraries, commercial libraries, …), you can now use Enamine’s HTS library, which contains 2M compounds in stock to our growing pool of prepared libraries. This will help you perform a virtual screening and purchase the selected compounds right away. If your preferred library is not yet in our pool, do not hesitate to let us know and we will assist you.
At Pharmacelera, we want to contribute to the open-source community. Because of that, we have updated our list of open-source solutions.
Events and Publications
We presented our technology at the GRC Computer-Aided Drug Design meeting in the US, the Computationally Driven Drug Design in Italy and the German Cheminformatics Conference. We have also attended several partnering and scientific events in the US and Europe. Visit our website for our forthcoming talks and events.
Regarding our scientific publications, we have a new article in the Journal of Future Medicinal Chemistry about the application of our hydrophobic descriptors to 3D-QSAR studies. We have a new article already prepared about the combination of docking protocols with our hydrophobic descriptors. Stay tuned for our next publications!
If you want to know more about Pharmacelera in 2020, just follow us on LinkedIn and Twitter!