Why protein-protein interactions are important?
Protein-protein interactions (PPI) play a major role in several of the cellular functions and processes that take place in our organism. Researchers estimate that around 130.000 binary interactions occur between different proteins (1). These interactions also regulate several of the pathogenic mechanisms that bacteria use during the infection process. The HPIDB (Human Pathogen Interaction Database) has registered close to 50.000 human-pathogen interactions (2). This fact and the overuse of traditional antibiotics, generating bacteria resistance, awake a great interest for protein-protein interaction inhibition.
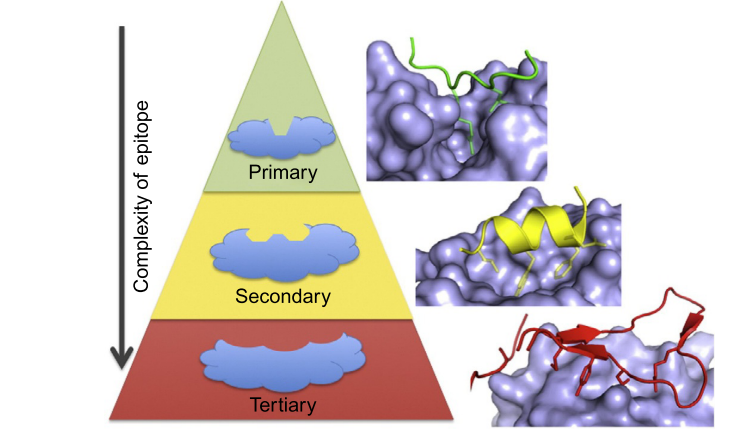
Protein-protein interactions are distinguished by flat and large surfaces when compared with the cavity-shaped binding pockets of an enzyme. Three classes of surfaces have been defined, based on the binders’ structural motifs: a peptide extended conformation that binds to a groove; single fold secondary structure, such as an alpha helix or beta barrel; and proteins with tertiary structures (3), associated with flatter surfaces. Traditionally, drug discovery programs based in disrupting protein-protein interactions have been largely de-prioritized due to the extended shape of the interacting surface. This trend changed years ago, thanks to the identification of hotspots (4,5), critical anchoring points for protein-protein recognition. These hotspots cluster in hydrophobic areas of the size of small molecules.
Approaches to find new inhibitors
The features of these binding surfaces and the presence of hotspots induced the generation of multiple approaches for PPI inhibition. One of the most promising is the fragment-based drug discovery (FBDD). Fragments are small molecules (< 250 Da) with binding affinities in the range of mM. They are excellent starting points for compound optimization, evolving to small molecules using growing, linking or merging strategies (6). Compared with small molecules, fragments detect new cavities and hotspots in protein surface due to their chemical diversity and small size. Nowadays, commercial vendors provide many different fragment libraries. And the number is still growing.
Peptidomimetics is a common approach when protein regions lead the PPI. In this case, linear or individual secondary structures bind to the target surface, driven by tens of residues. Following mutational studies and structural analysis, researchers can design compounds that mimic the main interactions between both proteins.
Macrocycles are a better approach when targeting flat protein surfaces. Macrocycles are ring-shaped molecules including cyclic peptides with a molecular weight in the range of 500-2000 Da. Macrocycle synthesis is still challenging. However, they retain small molecule properties, such as metabolic stability and the lack of immunogenicity. In addition, macrocycles show a binding affinity in the same range as antibodies. These properties make macrocycles very promising molecules (7).
More and more protein-protein crystal structures have been released, containing ligand (peptide or small molecule) binding information. New database resources (TIMBAL, Structure-PPi, 2P2I or PPI3D) complement known resources, such as PDB or Uniprot.
What can Pharmacelera do for you?
Results are showing that PPI inhibition is a promising but still challenging area of research. We need multiple approaches and methods to find the proper inhibitor due to the difficulty to target these surfaces. Although highly used, the majority of structure-based methods show difficulties in the identification of new hits considering the properties of these surfaces, mostly flat and hydrophobic. Conversely, ligand-based methods are protein agnostic approaches, offering an ideal option for PPI inhibitor development.
Following information obtained by known binders, our PharmScreen technology can be used to retrieve new potential molecules that would eventually inhibit PPI. PharmScreen is based on the usage of 3D hydrophobic molecular descriptors, which are ideal to describe the type of inhibitors that would bind to a protein-protein surface. Either peptidomimetic or fragment based strategies can be applied through PharmScreen.
Are you working on a PPI project? Let us help you! Our consultants will provide you with customized solutions and always working side by side with your team. Do you want to know more about our services and technologies? Contact us (contact@pharmacelera.com) for an open discussion with no commitment!
References
(1) Venkatesan, K., Rual, J.-F., Vazquez, A., Stelzl, U., Lemmens, I., Hirozane-Kishikawa, T. et al. (2009) An empirical framework for binary interactome mapping.Nat. Methods 6,83– 90
(2) Kumar, R., & Nanduri, B. (2010). HPIDB—A unified resource for host-pathogen interac- tions. BMC Bioinformatics, 11(6), S16.
(3) Arkin, M.R., Tang, Y. and Wells, J.A. (2014) Small-molecule inhibitors of protein–protein interactions: progressing toward the reality.Chem. Biol.21 , 1102– 1114
(4) Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995 Jan 20;267(5196):383-6.
(5) Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004 Apr;3(4):301-17. Review.
(6) Michèle N Schulz1 and Roderick E Hubbard “Recent progress in fragment-based lead discovery”, Current Opinion in Pharmacology 2009, 9:615–621
(7) Dougherty PG, Qian Z, Pei D. Macrocycles as protein-protein interaction inhibitors. Biochem J. 2017 Mar 15;474(7):1109-1125. Review.