Enric Herrero
May 30th, 2023
Commercial chemical libraries have witnessed remarkable growth in recent years, resulting in an unprecedented increase in size and diversity. With advancements in high-throughput synthesis and combinatorial chemistry techniques, compound providers like Enamine have expanded their collections of small organic molecules to meet the escalating demands of the pharmaceutical industry. This exponential growth has provided researchers worldwide with access to an extraordinary wealth of chemical diversity, facilitating the discovery and development of novel therapeutic agents.
However, the significant growth in size and diversity of commercial chemical libraries has rendered previous methods of virtual screening impractical, especially for accurate 3D methods. The sheer volume of compounds amassed within these libraries presents immense challenges in terms of storage and computational costs. Taking as a reference a compressed SD file containing multiple stereoisomers and conformers of a molecule of 48KB, the fully enumerated library of Enamine REAL (31 billion compounds) would require a storage capacity of 1.36 PB of data! This is almost 1400 hard drives like the one that you have in your laptop! Following the same example, if we assume that processing this single molecule requires 3.6 ms in your laptop, this will mean 3.5 years of calculations to perform a screening!
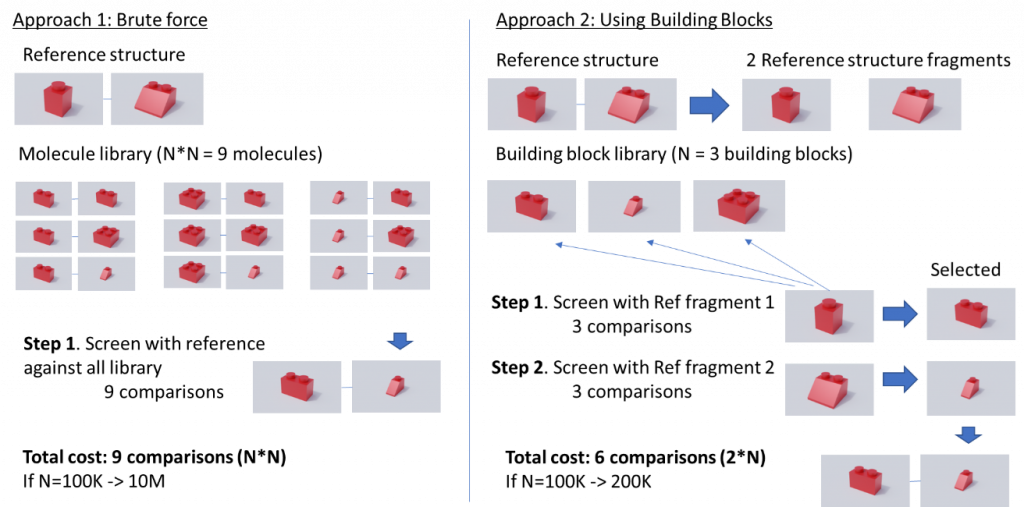
Luckily, several methods have been proposed that rely on the way these huge libraries are created, which is combining a set of building blocks to generate new molecules. Figure 1 shows an example of the value of using building blocks to perform a screening, for simplicity we will assume that each building block is a synthon and that all of them can be combined. In this example we have a building block library of 3 building blocks that can generate a chemical space of 9 molecules (combining all against all). If we apply the traditional approach (Brute force) we would compare our reference structure against each of the molecules of the library, this is 9 comparisons. However, if we perform a smart enumeration taking advantage of how this library has been created, we can reduce the computing cost. In this case what we would do is to partition the reference structure in two fragments and instead of comparing against all the enumerated library we perform the comparison against the building block library, this is 3 comparisons. Since we have two reference fragments we need to perform this operation twice, resulting in 6 comparisons, 3 less than in the brute force approach. This difference in the number of comparisons does not seem large but if we translate this example to a building block library of 100K building blocks, in the brute force approach we would need 10 million comparisons vs 200 thousand for the smart enumeration approach.
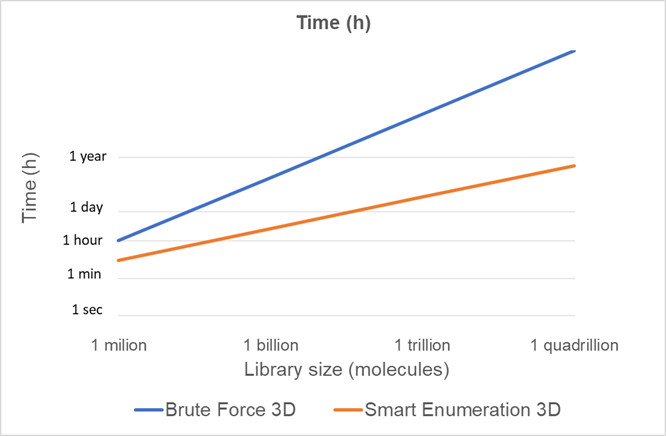
If we plot the computational cost projections of both methods for different library sizes (Figure 2) we can see how the scalability of the smart enumeration approach is much better than the brute force approach and, therefore, is much more suitable for the chemical libraries of the future.
Overall, we have seen that the rapid growth of commercial chemical libraries represents a challenge for virtual screening tools. The size and diversity of these libraries have made traditional screening methods impractical due to storage and computational costs. However, the use of smart enumeration based on building blocks offers a more efficient approach. By leveraging the way these libraries are created, researchers can significantly reduce the number of comparisons needed for screening. This smart enumeration approach shows better scalability and is considered more suitable for future chemical libraries, offering computational efficiency compared to brute force methods.